Mario Chiaparini
I'm
About

Hello There !
My name is Mario, I am a final-year Electrical Engineering student with a previous degree in Chemical Engineering and experience in scientific computing both in academia, at research centers, and in industry.
I began my career working at a bioinformatics company called Genera, which utilized genetic sequencing data for its products, including machine learning applications. At this company, I gained knowledge that attracted the interest of a scientific computing group at the Brazilian particle accelerator, Sirius, which is shown in the photo beside.
After two years of internship at Genera, I decided to accept the opportunity to work onsite at the Sirius particle accelerator on topics related to scientific computing, where I engaged in atomistic simulations (MD and DFT) and scientific machine learning with computer vision.
I worked with prominent figures in bioinformatics, such as Dr. Paulo Sergio Lopes and Dr. Helder Veiga, who greatly contributed to my knowledge in deep learning and computational modeling. It was during this time that I graduated in Chemical Engineering and continued with a degree in Electrical Engineering at the State University of Campinas.
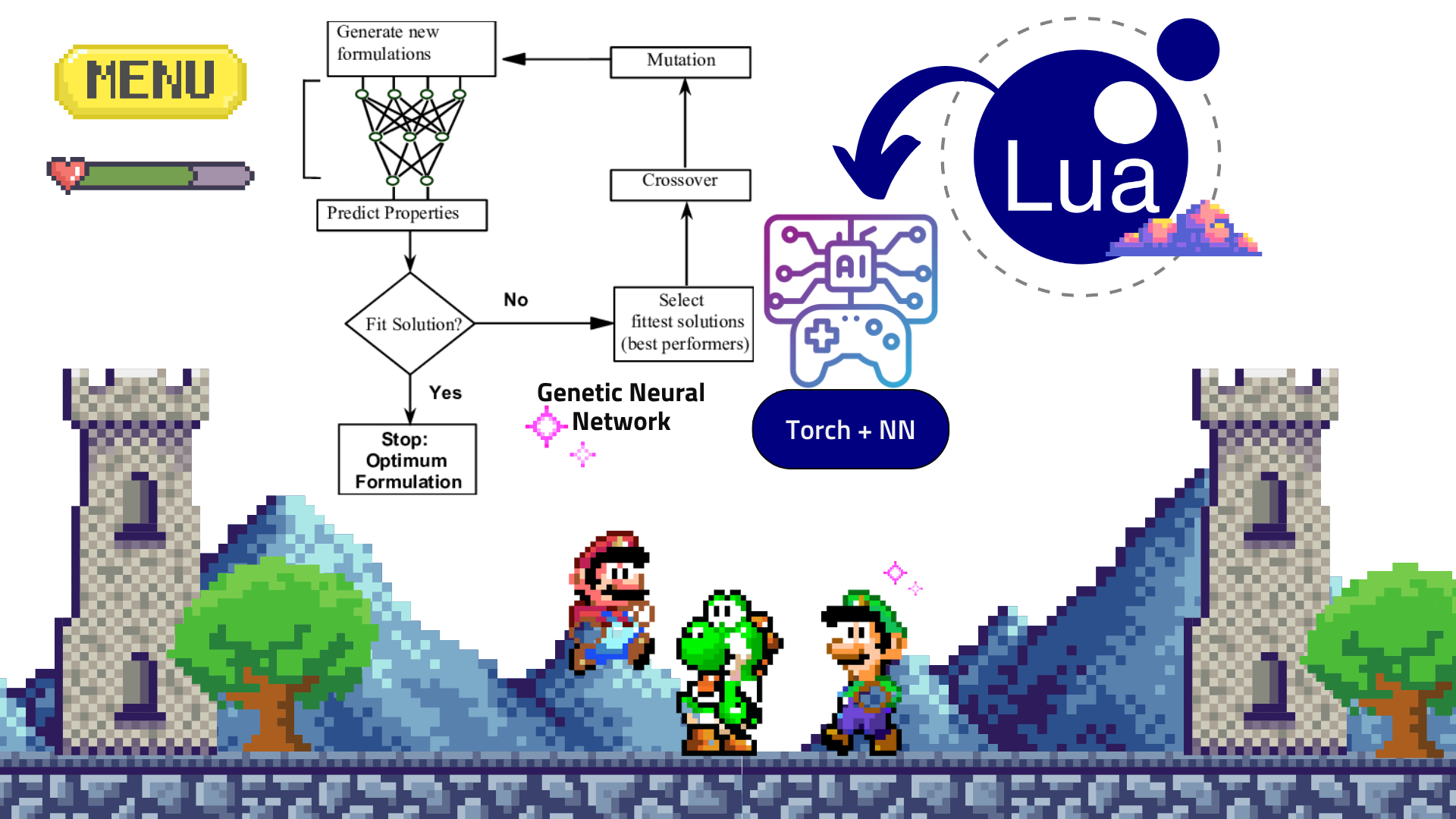
Upon completing my projects and configurations during my scientific computing internship, I was assigned to a research center called the Center for Innovation on New Energies, working with Professor Hudson Zannin's group on electrochemical simulations of capacitors and batteries. This work involved using Python and other computational tools, as well as applying data-driven methods like neural networks for battery data prediction.
I am currently finalizing this project in collaboration with TotalEnergies and starting my PhD in Quantum Computing with Professor Dr. Cristhian Rothenberg, focusing on the development of Qubit communication systems with photonics experiments and simulations, and utilizing quantum machine learning to optimize telecommunication systems computationally.
- Birthday: 29 September 1999
- Phone: +55 11945262188
- City: Indaiatuba, São Paulo
- Email: mariochiaparin@gmail.com
Interests
Computational Physics
Computer Vision
Deep Learning
Generative Models
Battery Informatics
Data analysis
Machine learning
Data Science
University

Universidade Estadual de Campinas
Eletrical Engineering
January 2023 - Present
Relevant Coursework
- Hardware Desing - PCB
- Circuit Analysis
- Eletromagnetism

Pontificia Universidade Católica
Chemical Engineering
January 2018 - December 2022
Relevant Coursework
- Mass Transfer
- Momentum Transfer
- Heat Transfer
Experience
Center for Innovation in New Energies, CINE
Jan 2023 - Present
Associate Researcher
- Worked at the Center for Innovation in New Energies, focusing on the battery informatics R&D team from Total Energies.
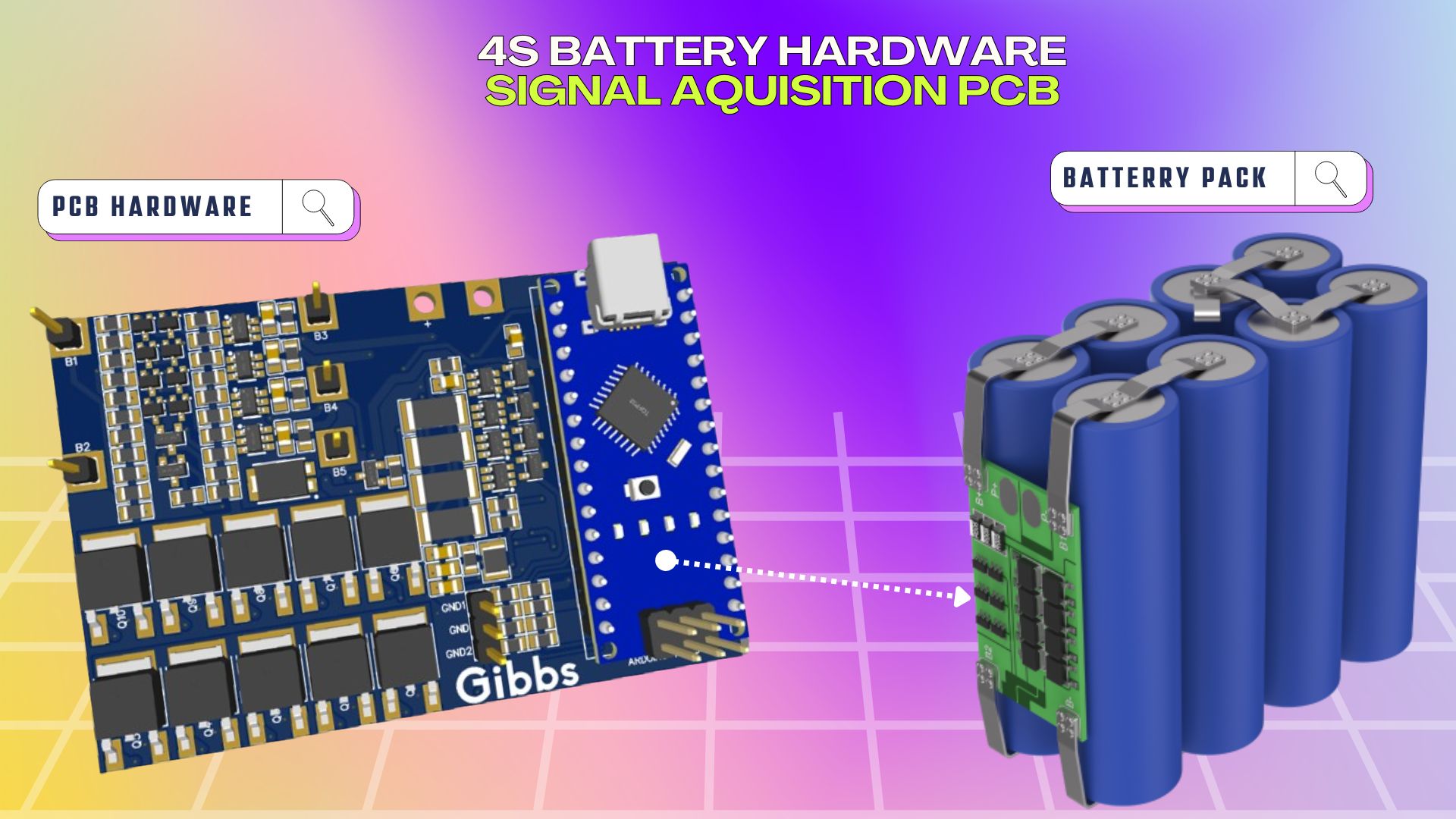
- Within the battery informatics segment, I worked in both hardware and software sections.
- In the software section, I worked on hardware development for battery management systems and signal acquisition.
- In addition to the hardware application for batteries, I have projects involving the development of supercapacitor chargers.
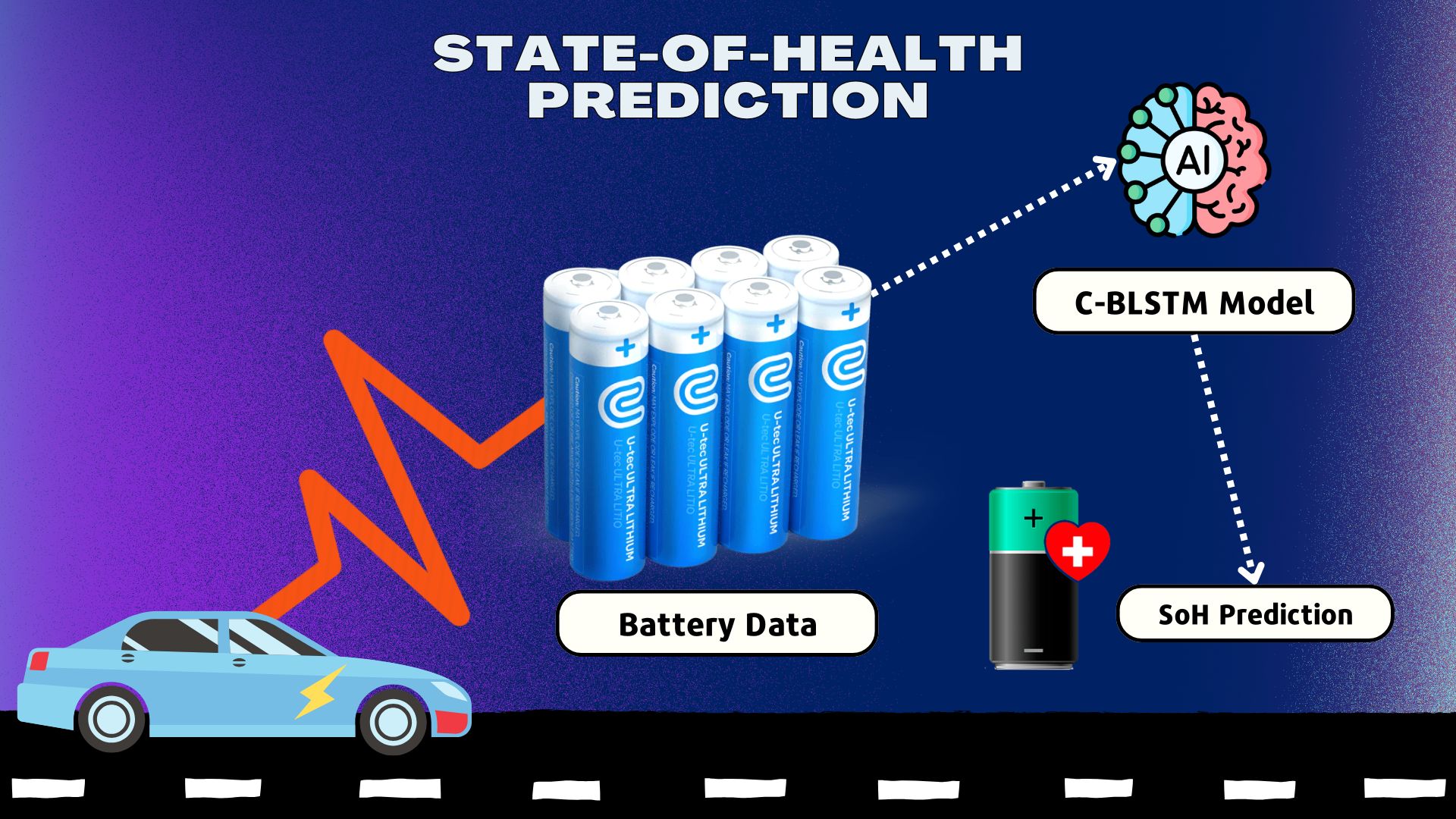
- Software development is primarily focused on signal processing, with the main application in health state prediction based on recurrent neural networks.
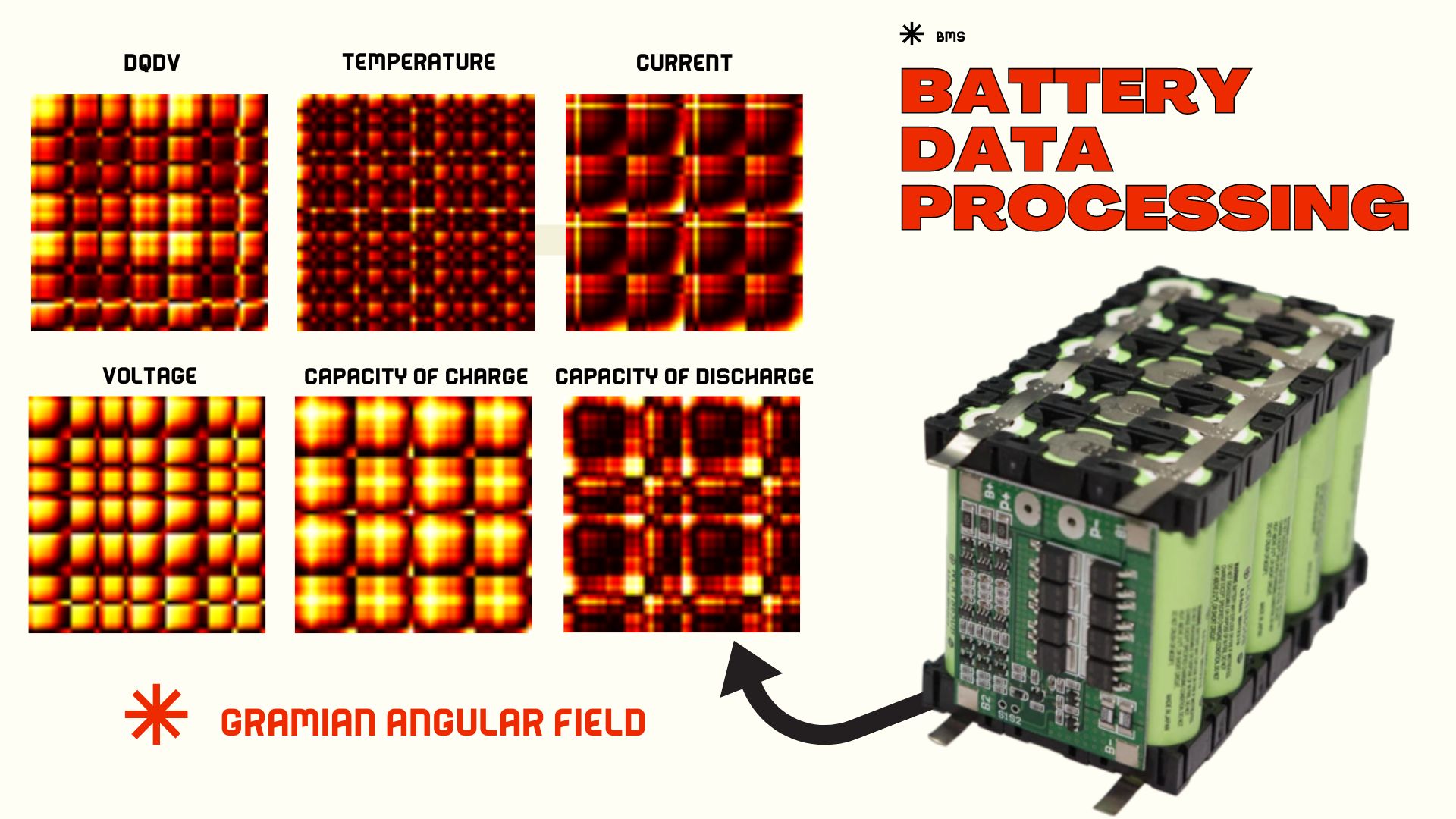
- In addition to health state prediction, I developed a convolutional network to define the health state up to the second life region of batteries, through a signal transformation.
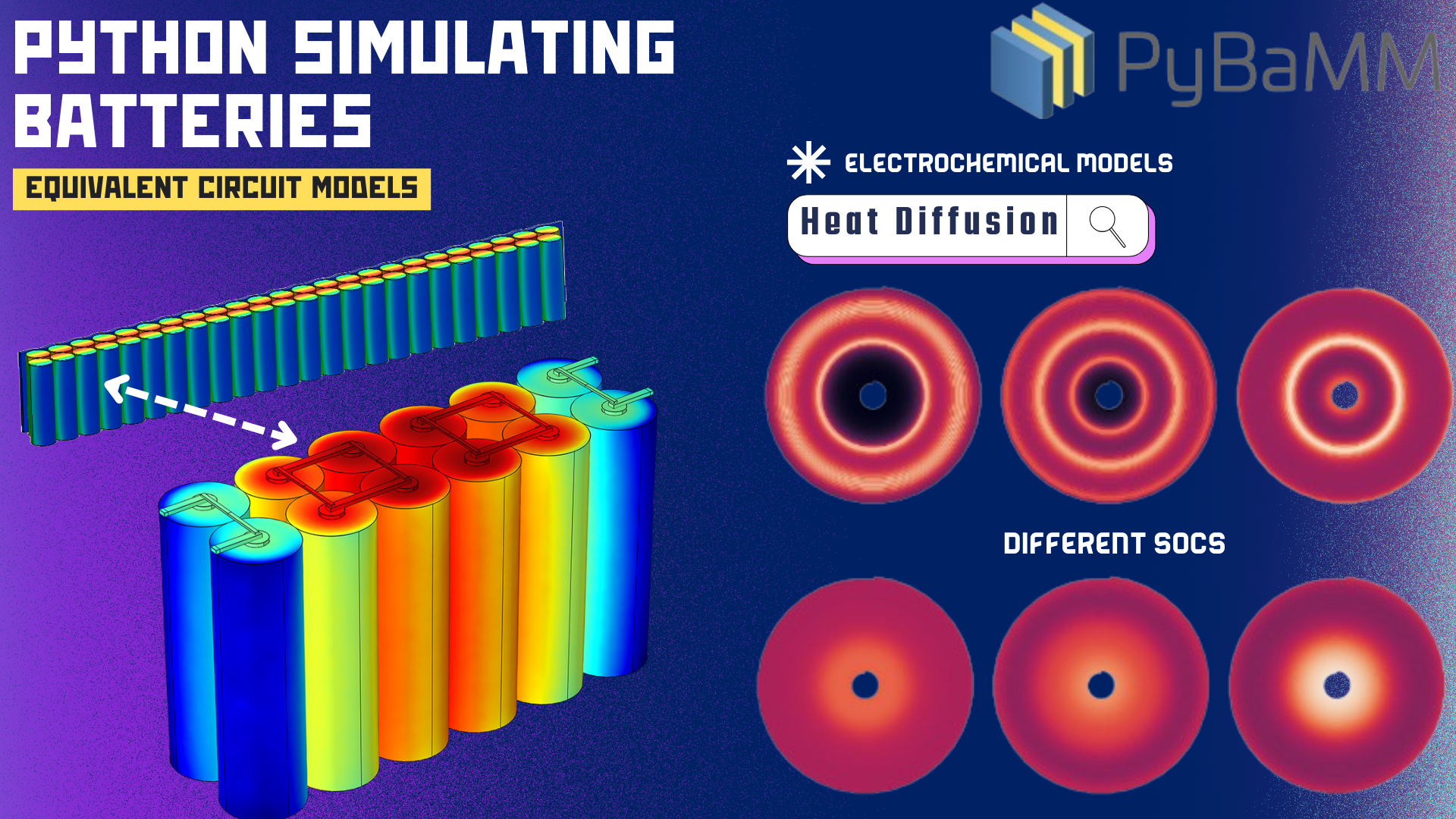
- In addition to data-driven methods applied to battery informatics, I developed a single-particle and Doyle-Fuller-Newman models that simulated the electrochemical systems from the batteries computationally.
Center of research in Materials and Energy, CNPEM
Jan 2022 - Jan 2023
Scientific computing intern
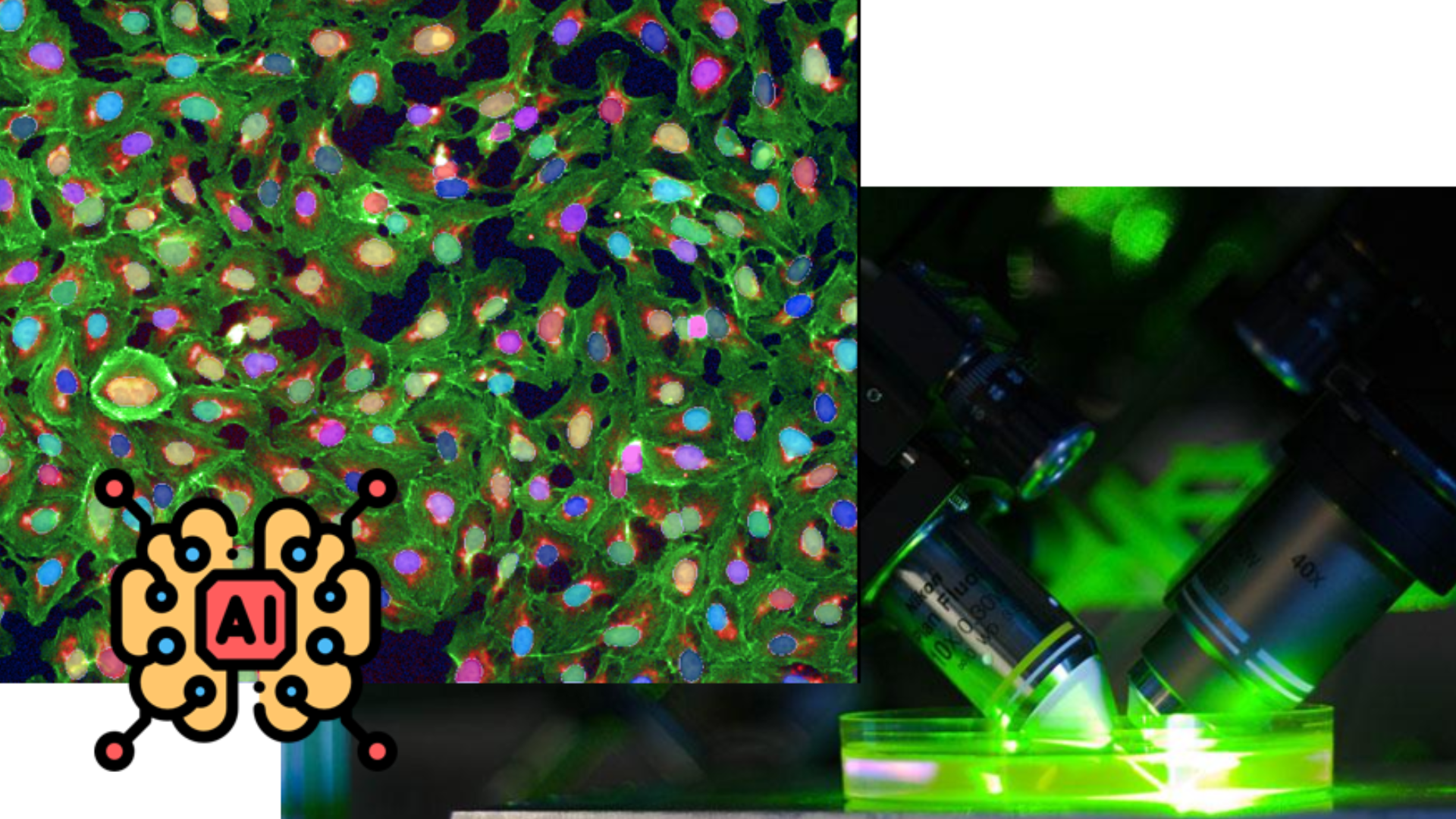
- Tested the convolutional neural networks (Deep learning models) to find and count patogenic microorganisms inside of synthetic human cells. The data was generated from high throughput screening experiments, to find new pharmaceuticals for Chagas disease.
- Deployed a deep learning model on a pipeline from Cellprofiler that have the exactly same functions as the lincesed software (Columbus), which saved 70 thousand dollars for the research group. Turning pharmaceutical discoverying, for new tropical diseases, cheapier to research in Brazil and any other south american country.
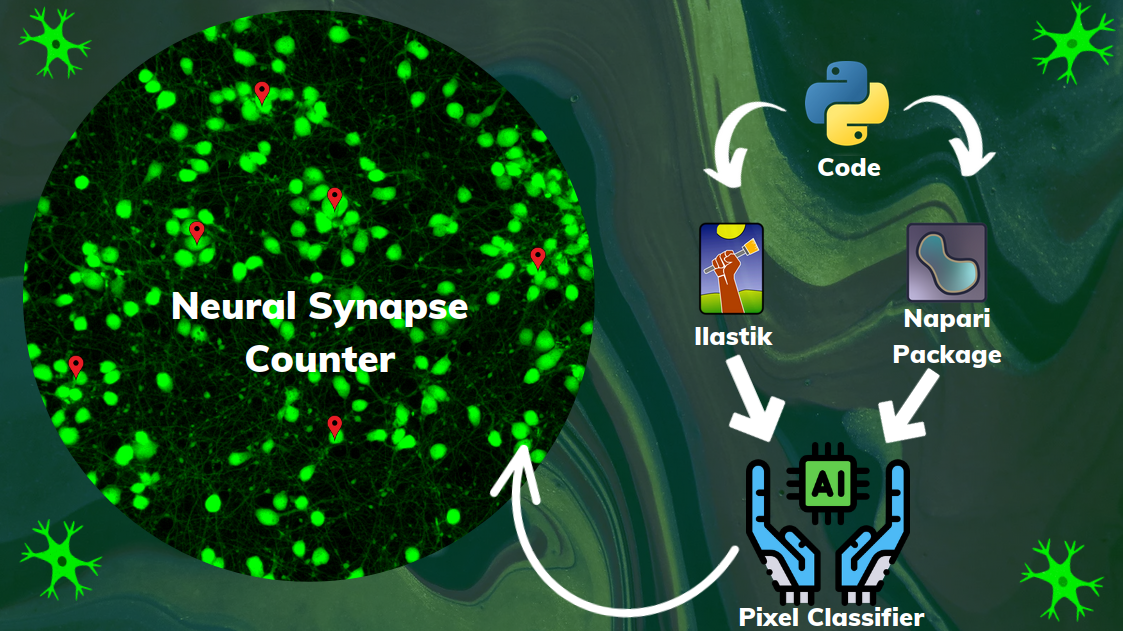
- I developed other convolutional neural networks which the input data was from synthetic human neural networks synapses.
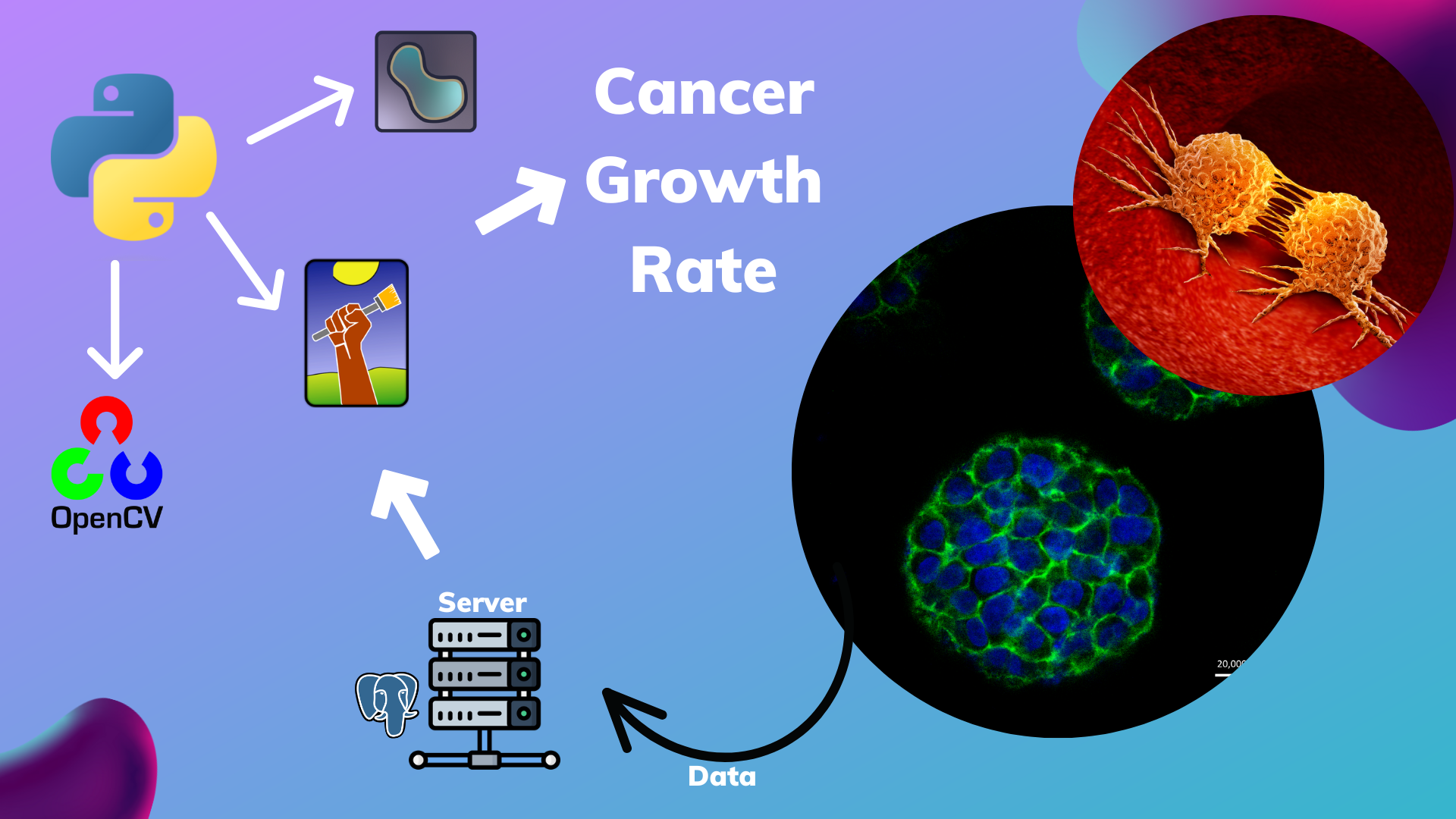
- Designed a pipeline with Cellprofiler and Ilastik segmentation model, that followed the tumorous cells growth. Saving the time for researchers, that studied hit molecules for all types of cancer cells.
- Developed python macros to perform molecular docking simulations, using the lincesed software called Yasara. I built these scripts to turn the process of molecular docking in Yasara easier.
- Built and tested Singularity and Docker images for molecular dynamics and molecular docking tools to be deployed on the HPC cluster.
- Developed scipts in bash for LAMMPS, Quantum Espresso and Gromacs, for the simulation data generated from the particle accelerator clusters.
- Handled with Ansible playbooks to install the enviroments to update each node, from the particle accelerator clusters of CPU and GPU.
- Worked with OpenCL, Mosix and Popcorn Linux to create an application for the syncrotron accelerator, a descentralized CPU cloud. Where I take every type of CPU on the particle accelerator, connected with each of the CPUs from the beamlines, with Mosix(OpenVCl) and VM interface created with Popcorn Linux
- Created the Linux clusters enviroment of processing, by installing Slurm for a parallel job execution and installing all the containers.The cluster named as "Tars" was for test application, which got only 3 nodes of CPU for some molecular models testing.
Genera Laboratories
Aug 2020 - Jan 2022
Data Science intern
- Bioinformatics tools for biological data analysis.
- Built a Hidden-Markov chain model to phase and impute the missing data from DNA, which imputed the SNPs from Illumina to thermoFisher.
- Tested bioinformatics tools for DNA data processing, with Plink and Lineage.
- Designed a pipeline to process the genetic data, to phase and impute the SNPS from each chip. Using the tools like MendelImpute.jl and Beagle, which was implemented on the AWS cloud.
- Developed a Python code to use VCTools and BCFTools to find genetic variants for the R&D group of the company.
- Dbeaver for database management
- Optimized the data processing from the DNA sequencing chips (Illumina vs ThermoFisher) to find "indels"
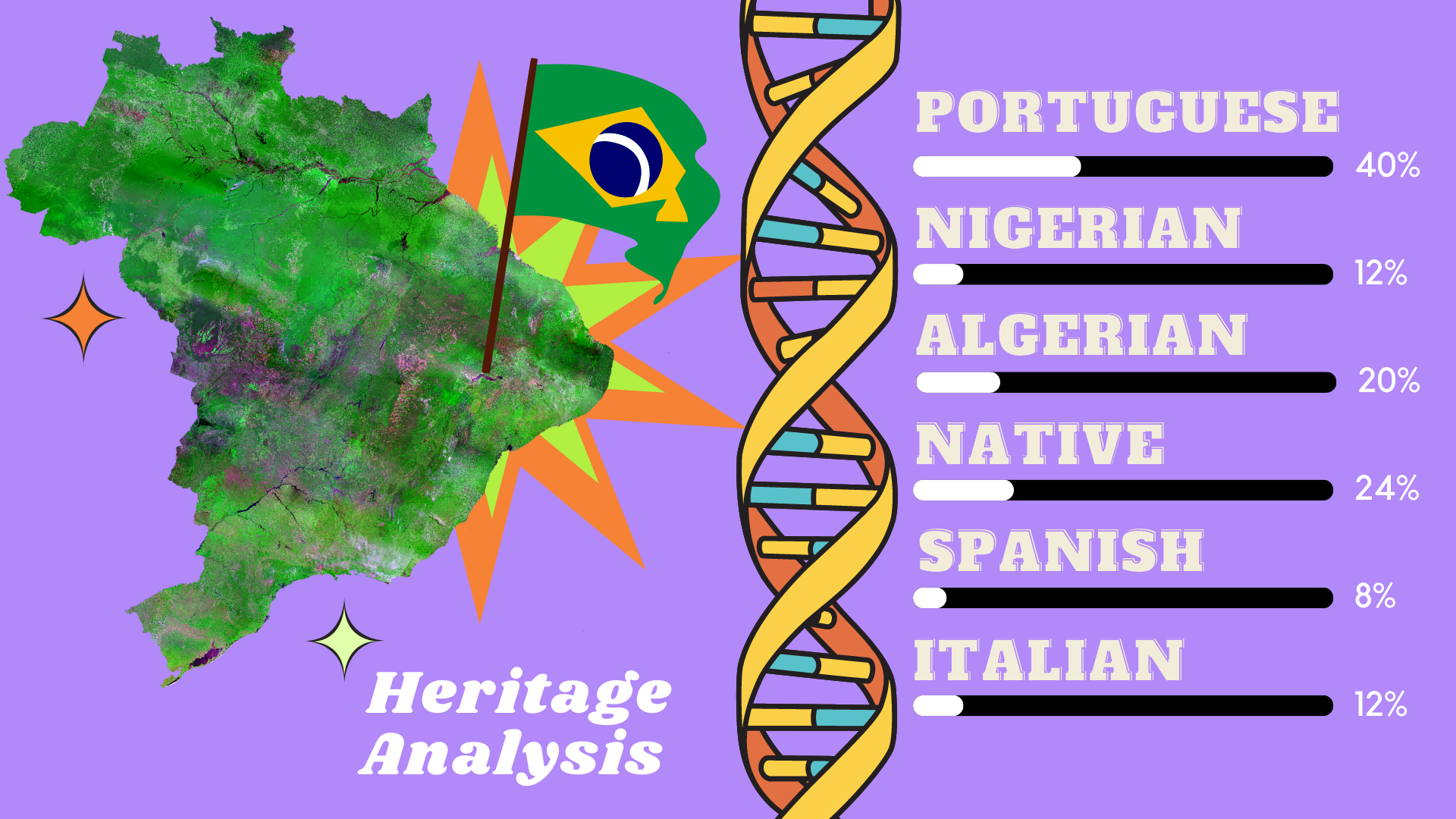
- Handled with the data analysis from the client's heritage, giving the ancestry map from brazilians by each province
Projects
- All
- Artificial Intelligence
- Computational Physics
- Other
Skills
Languages and Databases




Packages





![]()
![]()


![]()


Tools

![]()



![]()
![]()
![]()


![]()